Innovative molecular imaging reveals cellular metabolism
Cellular metabolism is defined by a complex interplay between molecules, molecular signals, and a dynamic environment. The detailed study of local metabolism provided insight into the heterogeneity of living systems from plant to human cells. Innovative single-cell imaging of complex surfaces, combined with advanced “omics” approaches, is employed to chart living systems in unique detail.

Focus Group: Molecular Imaging of Cellular Metabolism
Prof. Ron M. A. Heeren (University of Maastricht), Alumnus Hans Fischer Senior Fellow | Sutirtha Chattopadhyay (TUM University Hospital Klinikum rechts der Isar), Doctoral Candidate | Hosts: Prof. Thomas F. Hofmann (TUM), Prof. Percy A. Knolle (TUM University Hospital Klinikum rechts der Isar)
(Image: Astrid Eckert, TUM)
The objective of the Focus Group was three-fold. First to establish a close collaboration between the labs of Ron Heeren at the University of Maastricht and the research labs at TUM. The second, more scientific aim was to employ these technologies to characterize the metabolic profile of key immune cells and their interaction with mammalian cells. The third objective was to explore the tissue microenvironment in the direct vicinity of individual immune cells in liver and other tissues. Multimodal molecular imaging combined with spatial “omics” was the main technology used for the research conducted in this collaboration.
While in the early years of the Fellowship Covid-19 made certain topics more difficult to pursue, a new collaborative infrastructure for interdisciplinary research based on mass spectrometry imaging was established early on at TUM. This facilitated the scientific research the Focus Group aimed for. Several programs were set up to train TUM researchers at the Maastricht facilities, enabling the direct knowledge transfer needed to conduct research at TUM.
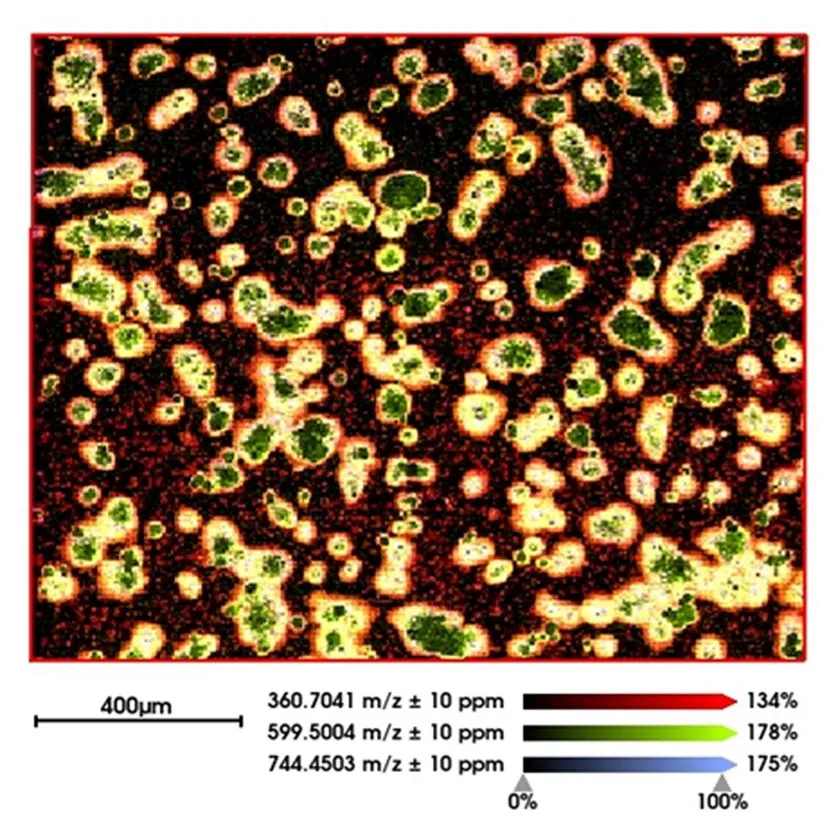
Initial research was aimed at establishing metabolic images of isolated primary immune cells with MALDI-MSI. In particular, we implemented and optimized post-ionization by MALDI-2 to overcome the effects of ion suppression and increase sensitivity. The increased sensitivity resulted in the improved spatial resolution required for single-cell metabolic imaging. This approach spawned an exciting new area of research for investigating the molecular causes and effects of cellular interactions. This was successfully realized and resulted in high-content, high-resolution images such as the molecular image of hepatocytes shown in Figure 1.
Figure 1
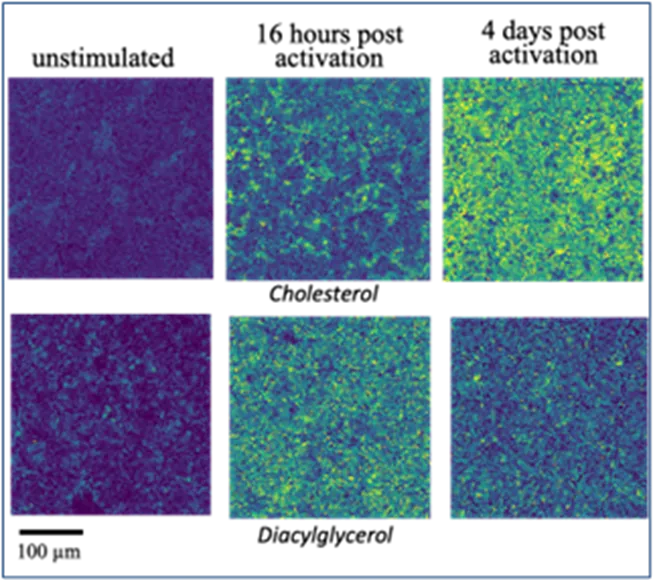
One of the experiments employed anti-CD3 activated CD8 T cells, the changing metabolic signature of which was determined through multimodal molecular imaging post-activation. The results demonstrated the feasibility of this approach to use this metabolic profile to determine the metabolic age of activated CD8 T cells (Figure 2). This identified metabolites and lipids as local mediators of cell-cell communication, a finding impossible to obtain with protein-based technologies. This formed the basis of several grant proposals submitted and the basis of future work on cell-cell communication. Moreover, the research combined targeted and untargeted approaches to build cell type-specific metabolome and lipidome signatures characteristic for specific functional states of immune cell populations in tissues. Machine learning-based analysis of the cell-cell communication between liver tissue cells and immune cells detected in situ are targeted to provide a new level of understanding of how immune responses are fine-tuned to the tissue microenvironment. This information has been demonstrated to be crucial in the understanding of virus-specific CD8 T cells in the liver. In a study published in Nature (2024), it was demonstrated that a liver immune rheostat renders these CD8 T cells refractory to activation and leads to their loss of effector functions.
Additional collaborative studies were conducted in close collaboration with Corinna Dawid on the spatial distribution of food- and flavor-related compounds in mushrooms and plant-based alternative meat products. The studies were based on a new protocol developed to generate metabolic images of plant-based samples and demonstrated the use of molecular imaging technologies in food and ingredient classification and authentication.
A close collaboration between Bastian Hoechst and the imaging team of M4i resulted in a better morphological understanding of breast cancer-isolated extracellular vesicles and their role in cell-cell communication. The work has been submitted for publication and is under review.
Figure 2
The findings of these and several other projects were used as the basis of a joint ERC-synergy application, in close collaboration with the Helmholtz Institute, with the target to establish a synergistic European research program in cell-cell communication in the immune system. Unfortunately, the program was not funded after the interview stage, but it did form the basis for future collaborations and proposals that take advantage of established network. TUM is a collaborator in a Dutch large-scale research infrastructure grant, BioBeyond_NL, and Ron Heeren is proposed as a Mercator Fellow in a DFG Collaborative Research Center, Dynamics and Determinants of Tissue Immunity, in which imaging MS will play a central role. Future research will entail the integration of spatial transcriptomics and spatial metabolomics using technologies under development at Maastricht University. Ron Heeren was named TUM Ambassador at the end of this Hans Fischer Senior Fellowship, which will facilitate strengthening the future collaborations on research and education in immune metabolism and molecular imaging.
Selected publications
- Vats, M., Flinders, B., Visvikis, T., Dawid, C., Hofmann, T. F., Cuypers, E. & Heeren, R. M. A. Mass spectrometry imaging for spatial ingredient classification in plant-based food. J. Am. Soc. Mass Spec., 36, 100-107 (2025).
- Bosch, M., Kallin, N., Donakonda, S., Zhang, J. D., Wintersteller, H. et al. A PKA-operated liver-tissue rheostat curbs T cell receptor signalling and effector function of virus-specific CD8 T cells in chronic viral hepatitis. Nature, 631, 867-875 (2024).
- Höchst, B., Wilhelm A., Flynn, C., Hammer, E., Roessler, J. et al. Analysis of plasma-derived extracellular vesicles to determine the HER2 status in breast cancer patients. Breast Cancer Res., submitted.
- Vats, M., Cillero-Pastor, B., Flinders, B., Cuypers, E. & Heeren, R. M. A. Mass spectrometry imaging protocol reveals flavor distribution in edible mushrooms. J. Food Sci. Technol., 61, 888-896 (2024).
- Dudek, M., Pfister, D., Donakonda, S., Filpe, P., Schneider, A. et al. Auto-aggressive CXCR6+ CD8 T cells cause liver immune pathology in NASH. Nature, 592, 444-449 (2021).