NMR-based structural biology of membrane proteins in a native lipid environment
The TUM-IAS Focus Group Structural Membrane Biochemistry works on the development of tools for the high-resolution structure determination of membrane proteins in a native lipid bilayer environment by nuclear magnetic resonance (NMR) spectroscopy and other structural methods. Since membrane proteins are very difficult to study with structural methods, advanced strategies are required.

Focus Group Structural Membrane Biochemistry
Prof. Franz Hagn (TUM), Alumnus Rudolf Mößbauer Tenure Track Professor | Dr. Inguna Goba (TUM), Postdoctoral Researcher | Melina Daniilidis, David Goricanec, Elisabeth Häusler, Kai Klöpfer, Kolio Raltchev, Laura Sperl, Andrea Steiner, (TUM), Doctoral Candidates | Host: Structural Membrane Biochemistry, TUM
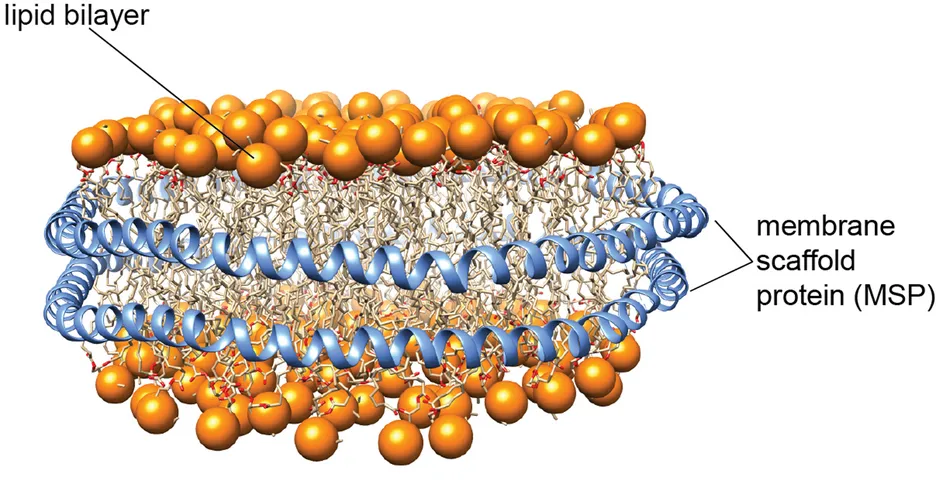
The development of native lipid bilayer mimetics is crucial to study sensitive membrane proteins and their complexes. In the last years, we were able to establish the use of so-called lipid nanodiscs for the investigation of the structure and dynamics of membrane proteins by solution-state NMR spectroscopy. Lipid nanodiscs consist of a patch of a lipid bilayer membrane encircled by two copies of a membrane scaffold protein (MSP) (Figure 1). To facilitate NMR and electron microscopy (EM) structural studies, we designed nanodiscs of different sizes, ranging from 6 to 26 nm in diameter [1]. To obtain better size homogeneity and stability of these nanoparticles, we applied protein ligation methods to produce circularized MSPs [2]. Furthermore, since sample homogeneity is essential for structural investigations, we developed a robust assay to probe and optimize the number of membrane proteins that are inserted into nanodiscs [3].
In addition to the development of lipid nanodiscs, we are dedicated to obtaining novel structural and functional insights on membrane proteins involved in signal transduction, apoptosis, and metabolite transport.
Figure 1
Membrane-anchored Bcl2 proteins play a major role in the regulation of apoptosis at the outer mitochondrial membrane. Pro-apoptotic members form pores or induce pore formation, leading to the release of pro-apoptotic factors. Anti-apoptotic members inhibit pore formation. We succeeded in producing these proteins in a lipid bilayer environment in high yields for structural studies by NMR [4] and were able to show that anti-apoptotic BclxL is located at the membrane surface, whereas pro-apoptotic, pore-forming members insert into the bilayer. This work will be the basis for studying these proteins and their complexes by structural methods in various states and in a lipid environment.
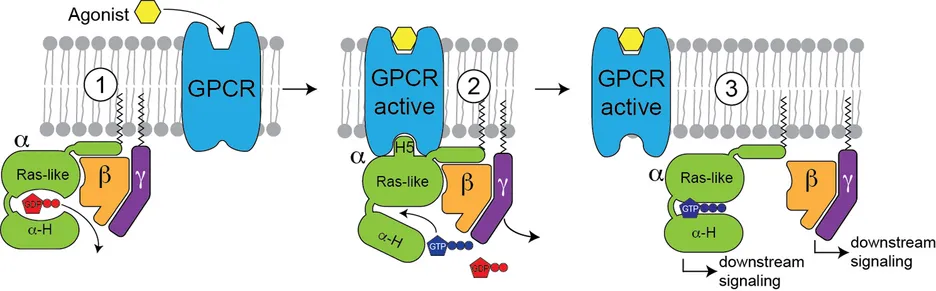
G-protein coupled receptors (GPCRs) and their associated G-proteins (Figure 2) represent another important membrane protein class where a native environment is crucial for functionality. GPCRs are very difficult to produce, especially for NMR. Using a stabilized GPCR (neurotensin receptor) obtained by directed evolution, we were able to study this important protein by NMR [5] and cryo-EM [6], and we were able to investigate the structural transition between the inactive and active states as well as the complex with a G-protein. Using NMR, we were also able to characterize the dynamical features a G-protein required for nucleotide exchange [7]. With these ongoing experiments, we will be able to define the structural hallmarks of GPCR activation, which will be of high interest for the design of novel small molecule inhibitors.
Figure 2
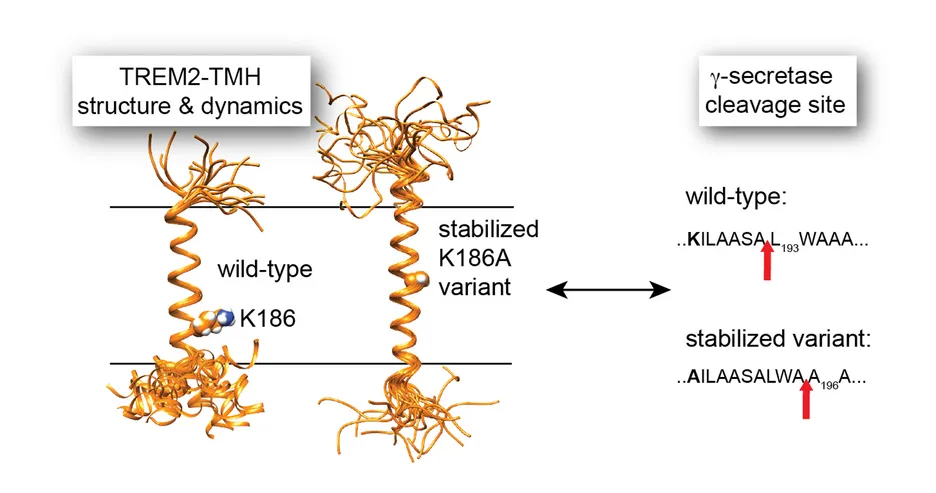
In a very recent study, we used NMR to determine the structure and functional features of the Alzheimer disease risk factor TREM2, a receptor on microglia cells in the brain that binds to Amyloid fibers and induces its clearance by phagocytosis [8]. The transmembrane domain of TREM2 is cleaved by the intramembrane protease γ-secretase. We show that the initial γ-secretase cleavage site is located in a dynamic region of TREM2. Disease-linked mutations in this region reduce its dynamics and lead to altered processing by γ-secretase. This study contributes to a better understanding of the substrate specificity of an intramembrane protease where recognition is based on structural and dynamical features (Figure 3).
With this toolset at hand, we are going to tackle challenging membrane protein systems in the future. A particular area of interest will be pore forming proteins as well as metabolite transporters in mammals and plants that are often difficult to characterize and consequently are only poorly explored.
Figure 3
[1]
F. Hagn, M. Etzkorn, T. Raschle and G. Wagner, “Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins”, Journal of the American Chemical Society, vol. 135, no. 5, pp. 1919-25, 2013; F. Hagn and G. Wagner, “Structure refinement and membrane positioning of selectively labeled OmpX in phospholipid nanodiscs”, Journal of Biomolecular NMR, vol. 61, no.3-4, pp. 249-60, 2015; F. Hagn, M.L. Nasr and G. Wagner, “Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR”, Nature Protocols, vol. 13, no. 1, 79-98, 2018; K. Klöpfer and F. Hagn, “Beyond detergent micelles: The advantages and applications of non-micellar and lipid-based membrane mimetics for solution-state NMR”, Progress in Nuclear Magnetic Resonance Spectroscopy, vol. 114-115, pp. 271-283, 2019.
[2]
M. L. Nasr et al., “Covalently circularized nanodiscs for studying membrane proteins and viral entry”, Nature Methods, vol. 14, no. 1, pp. 49-52, 2017; J. Miehling, D. Goricanec and F. Hagn, “A Split-Intein-Based Method for the Efficient Production of Circularized Nanodiscs for Structural Studies of Membrane Proteins”, ChemBioChem, vol. 19, no. 18, pp. 1927-1933, 2018.
[3]
E. Häusler et al., “Quantifying the insertion of membrane proteins into lipid bilayer nanodiscs using a fusion protein strategy”, Biochimica et Biophysica Acta - Biomembranes, vol. 1862, no. 4, pp. 183190, 2020.
[4]
K. Raltchev, J. Pipercevic and F. Hagn, “Production and Structural Analysis of Membrane-Anchored Proteins in Phospholipid Nanodiscs”, Chemistry, vol. 24, no. 21, pp. 5493-5499, 2018.
[5]
I. Goba et al., “Probing the Conformation States of Neurotensin Receptor 1 Variants by NMR Site-Directed Methyl Labeling”, ChemBioChem, vol. 22, no. 1, pp. 139-146, 2021.
[6]
M. Zhang et al., “Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs”, Nature Structural & Molecular Biology, vol. 28, no. 3, pp. 258-267, 2021.
[7]
D. Goricanec and F. Hagn, “NMR backbone and methyl resonance assignments of an inhibitory G-alpha subunit in complex with GDP”, Biomolecular NMR Assignments, vol. 13, no. 1, pp. 131-137, 2019; D. Goricanec et al., “Conformational dynamics of a G-protein alpha subunit is tightly regulated by nucleotide binding”, Proceedings of the National Academy of Sciences of the United States of America, vol. 113, no. 26, pp. E3629-38, 2016; B. Zeng et al., “Structure, Function, and Dynamics of the Galpha Binding Domain of Ric-8A”, Structure, vol. 27, no.7, pp. 1137-1147 e5, 2019.
[8]
A. Steiner et al., “g-Secretase cleavage of the Alzheimer risk factor TREM2 is determined by its intrinsic structural dynamics”, The EMBO Journal, vol. 39, no. 20, pp. e104247, 2020.