Carbon catabolite repression in filamentous fungi
Filamentous fungi are important for the production of lignocellulolytic enzymes. Carbon catabolite repression (CCR) plays an important role in this process. We studied CCR in two of the most important reference fungi, Aspergillus nidulans and Neurospora crassa. We investigated the regulation of the transcription factor CreA in CCR and the role played by F-box proteins during the CCR regulation.


Focus Group: Fungal Sensing and Signaling of the Environment
Prof. Gustavo H. Goldman (University of São Paulo), Alumnus Hans Fischer Senior Fellow | Lisa Meyer (TUM), Doctoral Candidate | Host: Prof. J. Philipp Benz (TUM)
(Image: Andreas Heddergott (Benz))
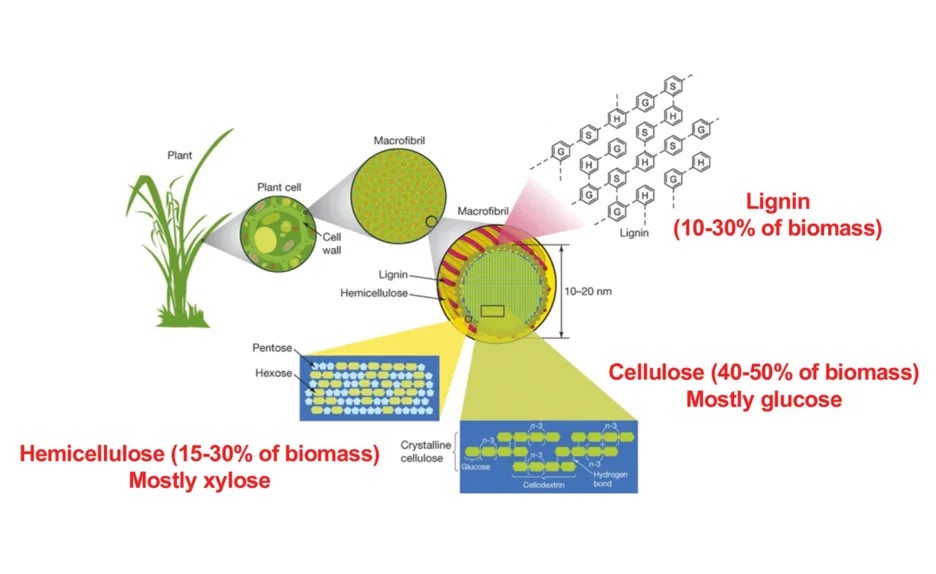
Filamentous fungi are of particular interest for biotechnological applications due to their natural capacity to secrete carbohydrate-active enzymes (CAZy) that target plant biomass (Fig. 1). The presence of easily metabolizable sugars, such as glucose, whose concentrations increase during plant biomass hydrolysis, results in the repression of CAZy-encoding genes in a process known as carbon catabolite repression (CCR), which is undesired for the purpose of large-scale enzyme production. Glucose is the preferential carbon source for most microorganisms, because it is rapidly metabolized, generating quick energy for growth. In the filamentous fungi A. nidulans and N. crassa, CCR is mediated by the transcription factor CreA / CRE-1, a C2H2 finger domain DNA-binding protein [1] (Fig. 2). The main aims of our project were: (i) to understand the regulation of the transcription factor CreA / CRE-1 during CCR and (ii) to investigate the role played by F-box proteins during the regulation of CCR. Our project is unique in the field, considering that, in investigating the regulation of this process, we are looking at both transcriptional and post-translational regulation.
CreA has been described as the major carbon catabolite repressor in Aspergillus spp., although little is known about the role of post-translational modifications in this process. We have identified S262, S319, S268, and T308 as phosphorylation sites. Sites S262, S268, and T308 are important for CreA protein accumulation and cellular localization, DNA binding, and repression of enzyme activities [2]. Site S319 was not important for several here-tested phenotypes but is key for CreA degradation and induction of enzyme activities. All sites were shown to be important for glycogen and Wtrehalose metabolism [2].
The utilization of different carbon sources in filamentous fungi underlies a complex regulatory network governed by signaling events of different protein kinase pathways. An initial screening of a library of 103 A. nidulans non-essential protein kinase (NPK) deletion strains identified several mitogen-activated protein kinases (MAPKs) to be important for CCR [3]. We selected the MAPKs Ste7, MpkB, and PbsA for further characterization and showed that they are pivotal for HOG pathway activation, protein kinase A activity, and CCR via regulation of CreA cellular localization and protein accumulation, as well as for hydrolytic enzyme secretion. Protein-protein interaction studies show that Ste7, MpkB, and PbsA are part of the same protein complex that regulates CreA cellular localization in the presence of xylan and that this complex dissociates upon the addition of glucose, thus allowing CCR to proceed [3]. Glycogen synthase kinase (GSK) A was also identified as part of this protein complex and shown to potentially phosphorylate two serine residues of the HOG MAPKK PbsA [3].
The attachment of one or more ubiquitin molecules by SCF (Skp-Cullin-F-box) complexes to protein substrates targets them for subsequent degradation by the 26S proteasome, allowing the control of numerous cellular processes. Glucose-mediated signaling and subsequent CCR are processes relying on the functional regulation of target proteins, ultimately controlling the utilization of this carbon source. There are 74 genes encoding F-box proteins in A. nidulans [4]. The Fbx23 protein was identified as being involved in CCR and the Δfbx23 mutant presented impaired xylanase production under repressing (glucose) and derepressing (xylan) conditions [5, 2]. Mass spectrometry showed that Fbx23 is part of an SCF ubiquitin ligase complex that is bridged via the GskA protein kinase to the CreA-SsnF-RcoA repressor complex, resulting in the degradation of the latter under derepressing conditions [5, 2]. Upon the addition of glucose, CreA dissociates from the ubiquitin ligase complex and is transported into the nucleus. Furthermore, casein kinase is important for CreA function during glucose signaling, although the exact role of phosphorylation in CCR remains to be determined [5, 2].
Figure 1
The attachment of one or more ubiquitin molecules by SCF (Skp-Cullin-F-box) complexes to protein substrates targets them for subsequent degradation by the 26S proteasome, allowing the control of numerous cellular processes. Glucose-mediated signaling and subsequent CCR are processes relying on the functional regulation of target proteins, ultimately controlling the utilization of this carbon source. There are 74 genes encoding F-box proteins in A. nidulans [4]. The Fbx23 protein was identified as being involved in CCR and the Δfbx23 mutant presented impaired xylanase production under repressing (glucose) and derepressing (xylan) conditions [5, 2]. Mass spectrometry showed that Fbx23 is part of an SCF ubiquitin ligase complex that is bridged via the GskA protein kinase to the CreA-SsnF-RcoA repressor complex, resulting in the degradation of the latter under derepressing conditions [5, 2]. Upon the addition of glucose, CreA dissociates from the ubiquitin ligase complex and is transported into the nucleus. Furthermore, casein kinase is important for CreA function during glucose signaling, although the exact role of phosphorylation in CCR remains to be determined [5, 2].
The main conclusions of our work are:
(i) Several novel aspects of the post-translational regulation of A. nidulans and N. crassa CreA / CRE1 and CCR were identified, highlighting for example the importance of CreA phosphorylation for the regulation of CCR.
(ii) Our work provides a model where phosphorylation events and the correct integration of PKA, HOG, and GSK signaling are required for the utilization of different carbon sources.
(iii) Finally, our screen of F-box-encoding genes demonstrates how targeted protein degradation is necessary for the fungal ability to switch between metabolic conditions.
These CreA / CRE-1 sites are interesting targets for biotechnological strain engineering without the need to delete essential genes, which could result in undesired side effects. These novel functions serve as a basis for additional research in fungal carbon metabolism with the aim of potentially improving fungal industrial applications. Future work will address the possibility of constructing genetically improved strains for increased enzymatic production.
Figure 2
[1]
Brown, N. A., Ries, L. N. A., Goldman, G.H. How nutritional status signalling coordinates metabolism and lignocellulolytic enzyme secretion. Fungal Genet. Biol. 72, 48-63 (2014).
[2]
de Assis, L. J. et al. (2021).
[3]
de Assis, L. J. et al. (2020).
[4]
Sarikaya Bayram, Ö. et al. (2022).
[5]
de Assis, L. J. et al. Regulation of Aspergillus nidulans CreA-Mediated Catabolite Repression by the F-Box Proteins Fbx23 and Fbx47. mBio. 9, e00840-18 (2018).
Selected publications
-
Horta, M. A. C. et al. Broad Substrate-Specific Phosphorylation Events Are Associated With the Initial Stage of Plant Cell Wall Recognition in Neurospora crassa. Front. Microbiol. 10, 2317 (2019).
-
de Assis, L. J. et al. The High Osmolarity Glycerol Mitogen-Activated Protein Kinase regulates glucose catabolite repression in filamentous fungi. PLoS Genet. 16, e1008996 (2020).
-
Gabriel, R. et al. The F-box protein gene exo-1 is a target for reverse engineering enzyme hypersecretion in filamentous fungi. Proc. Natl. Acad. Sci. USA 118, e2025689118 (2021).
-
de Assis, L. J. et al. Carbon Catabolite Repression in Filamentous Fungi Is Regulated by Phosphorylation of the Transcription Factor CreA. mBio. 12, e03146-20 (2021).
-
Sarikaya Bayram, O. et al. F-box receptor mediated control of substrate stability and subcellular location organizes cellular development of Aspergillus nidulans. PLoS Genet. 18, e1010502 (2022).