Expanding the genetic code: novel chemistries in living systems
The Focus Group for Synthetic Biochemistry was part of the TUM-IAS family from April 2014 until April 2020, during which time I was a Rudolf Mößbauer Fellow at the TUM-IAS and appointed as a W2 Tenure Track Assistant Professor in the Department of Chemistry at TUM. In April 2020 I was tenured as a W3 Associate Professor at TUM, and since April 2021 I am Full Professor for Chemical Biology at ETH Zurich, Switzerland.

Focus Group Synthetic Biochemistry
Prof. Kathrin Lang (TUM), Alumna Rudolf Mößbauer Tenure Track Professor | Marko Cigler, Maximilian Fottner, Marie-Lena Jokisch, Kristina Krauskopf, Susanne Mayer, Anton Murnauer, Marie-Kristin von Wrisberg, Vera Wanka, (TUM), Doctoral Candidates | Host: Synthetic Biochemistry, TUM
Our lab's research interests lie in the fascinating interdisciplinary area between chemistry and biology, applying concepts from organic chemistry and protein engineering to study and manipulate biological systems. Very much in the spirit of Richard P. Feynman, who coined the saying “What I cannot create, I do not understand,” we are convinced that an interdisciplinary approach between chemistry and biology is not only uniquely placed to increase our understanding of biological processes by developing tools to control biological activities, but is also indispensable for designing novel artificial biological pathways and ultimately even for engineering artificial living systems.
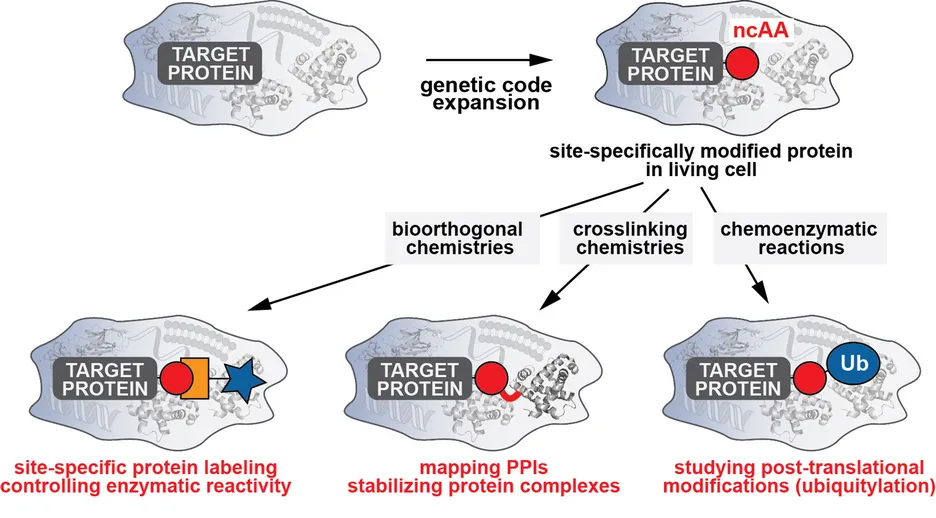
Our group has been especially active in developing and advancing approaches to endow biomolecules with new chemical reactivities within their physiological environment [1, 2]. In contrast to more traditional approaches of organic chemistry, our focus lies therefore not on creating ever more complex reactions in defined, laboratory settings, but on striving to discover novel reactivities that are amenable to and selective within the most complex environment: the living cell. In particular, we are interested in enabling and promoting methods to expand the genetic code by introducing artificial non-canonical amino acids (ncAAs) into proteins and in developing new in vivo chemistries to endow proteins with new chemical moieties, a combination that is ideally suited to addressing unmet challenges in studying and manipulating biological processes [2, 3].
Nature uses a limited set of 20 amino acids to synthesize proteins. In recent years it has become possible to genetically encode an expanded set of designer amino acids with tailored chemical and physical properties into proteins in bacteria and eukaryotes by reprogramming the genetic code and rewiring the translational machinery [3]. These strategies have started to have a big impact on biological studies, as they enable diverse applications including approaches for imaging and probing proteins, controlling and manipulating protein activity, and engineering and designing new protein functions and protein therapeutics.
My group has contributed over the last seven years to this exciting endeavor and has developed new tools for investigating and modulating molecular networks in living cells. We take a multidisciplinary approach and combine organic chemical, biochemical, biophysical, structural, and cell biological techniques to unravel the complex world of biology.
In my time as a Rudolf Mößbauer Fellow at the TUM-IAS and Professor at TUM, I have developed a vigorous research program to expand the genetic code and endow proteins with new functionalities in living cells. To expand the chemical space and functional repertoire of proteins within living cells, we have developed and applied rational and evolutionary approaches aimed at engineering components of the protein translational machinery that enable the site-specific incorporation of custom-designed ncAAs acids into proteins in living cells. By equipping proteins with functionalities beyond the ones provided by the 20 natural amino acids, we have contributed to three main research areas:
Figure 1
1
Bioorthogonal chemistries for studying and controlling protein activity: We have developed approaches to site-specifically endow proteins with ncAAs bearing bioorthogonal handles that allow the selective and rapid functionalization of proteins within living cells under physiological conditions. This has enabled us not only to image and probe proteins in vivo with various biophysical probes, but also to control and manipulate their enzymatic activity within their physiological environment [1] [4-11].
2
Chemical tools to validate and manipulate protein-protein interactions: A second research line in our lab involves development of crosslinking chemistries to study and validate protein-protein interactions (PPIs). PPIs are central to many biological processes. A considerable challenge, however, consists in understanding and deciphering when and how proteins interact, and this can be particularly difficult when interactions are weak and transient. In recent years we have developed approaches to site-specifically install ncAAs that upon a certain trigger (proximity or light) form covalent interactions with proteins in their vicinity. This allows us to covalently trap PPIs and thereby identify novel interactors, and to stabilize low-affinity and transient protein complexes to pioneer their structural elucidations [12-15].
3
Novel tools to generate and study post-translational modifications: In a third research line we are pioneering approaches in which site-specifically introduced ncAAs serve as a platform for tailored, chemoenzymatic reactions to site-specifically ubiquitylate target proteins both in vitro and in vivo [16]. Ubiquitylation represents a post-translational modification that controls almost every process in eukaryotic cells and has as such attracted considerable interest and promise as a therapeutic target. We have very recently been awarded an ERC Consolidator Grant to further develop an interdisciplinary chemical and synthetic biology platform that will provide unique insight into ubiquitylation and will lay the foundation for identifying new drug targets within the ubiquitin system.
Looking ahead to the future, our aims lie in understanding mechanisms of complex biological processes such as ubiquitylation through the application of synthetic molecules with tailored functions and properties. In particular, we plan to extend approaches for endowing proteins with new chemical moieties and thereby re-engineering and designing new protein functions. This will open up many possibilities for synthetic biology, drug design, biomaterials, and gene therapy.
[1]
K. Lang and J. W. Chin, “Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins”, Chemical Reviews, vol. 114, no. 9, pp. 4764-806, 2014.
[2]
K. Krauskopf and K. Lang, “Increasing the chemical space of proteins in living cells via genetic code expansion”, Current Opinion in Chemical Biology, vol. 58, pp. 112-120, 2020.
[3]
J. W. Chin, “Expanding and reprogramming the genetic code”, Nature, vol. 550, no. 7674, pp. 53-60, 2017.
[4]
K. Lang and J. W. Chin, “Bioorthogonal reactions for labeling proteins”, ACS Chemical Biology, vol. 9, no. 1, pp. 16-20, 2014.
[5]
K. Lang et al., “Genetic Encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions”, Journal of the American Chemical Society, vol. 134, no. 25, pp. 10317-10320, 2012.
[6]
K. Lang et al., “Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction”, Nature Chemistry, vol. 4, no. 4, pp. 298-304, 2012.
[7]
T. S. Elliott et al., “Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal”, Nature Biotechnology, vol. 32, no. 5, pp. 465-72, 2014.
[8]
C. Uttamapinant et al., “Genetic code expansion enables live-cell and super-resolution imaging of site-specifically labeled cellular proteins”, Journal of the American Chemical Society, vol.137, no. 14, pp. 4602-4605, 2015.
[9]
Y. H. Tsai, S. Essig, J. R. James, K. Lang and J. W. Chin, “Selective, rapid and optically switchable regulation of protein function in live mammalian cells”, Nature Chemistry, vol. 7, pp. 554-561, 2015.
[10]
S. Mayer, S. and K. Lang, “Tetrazines in Inverse-Electron-Demand Diels–Alder Cycloadditions and Their Use in Biology”, Synthesis, vol. 49, no. 4, pp. 830-848, 2017.
[11]
S. V. Mayer, A. Murnauer, M. K. von Wrisberg, M. L. Jokisch and K. Lang, “Photo-Induced and Rapid Labeling of Tetrazine-Bearing Proteins via Cyclopropenone-Caged Bicyclononynes”, Angewandte Chemie International Edition in English, vol. 58, no. 44, pp. 15876-15882, 2019.
[12]
T. A. Nguyen, M. Cigler and K. Lang, “Expanding the Genetic Code to Study Protein-Protein Interactions”, Angewandte Chemie International Edition in English, vol. 57, no. 44, pp. 14350-14361, 2018.
[13]
M. Cigler et al., “Proximity-Triggered Covalent Stabilization of Low-Affinity Protein Complexes In Vitro and In Vivo”, Angewandte Chemie International Edition in English, vol. 56, no. 49, pp. 15737-15741, 2017.
[14]
S. Tremel et al., “Structural basis for VPS34 kinase activation by Rab1 and Rab5 on membranes”, Nature Communications, vol. 12, no. 1, 1564, 2021.
[15]
J. Du et al., “Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1”, Nature Communications, vol. 12, no. 1, 460, 2021.
[16]
M. Fottner et al., “Site-specific ubiquitylation and SUMOylation using genetic-code expansion and sortase”, Nature Chemical Biology, vol. 15, no. 3, pp. 276-284, 2019.