How cells make their proteins – from insights to engineering
This is the final annual report from our Focus Group for Cellular Protein Biochemistry at the TUM-IAS. Matthias Feige, heading the group as a Rudolf Mößbauer Tenure Track Assistant Professor, has just obtained tenure and now continues his career as Associate Professor for Cellular Protein Biochemistry at TUM. This provides the opportunity to summarize some our achievements during the Fellowship as well to provide a few glimpses of the path ahead.

Focus Group Cellular Protein Biochemistry
Prof. Matthias J. Feige (TUM), Alumnus Rudolf Mößbauer Tenure Track Professor | Nicolas Blömeke, Joao Coelho, Karen Hildenbrand, Susanne Meier, Yonatan Mideksa, Stephanie Müller, (TUM), Doctoral Candidates | Host: Cellular Protein Biochemistry, TUM
In complex organisms, cells need to communicate and interact with each other. This communication is the basis of growth, development, memory formation, and immune defense. Key players in all cellular communication processes are proteins secreted from cells or displayed on the cell surface. Within every mammalian cell, like our human cells, a specialized sub-compartment exists that is responsible for the production of these proteins: the endoplasmic reticulum (ER). Its name stems from its microscopic appearance: a net-like structure within the interior of the cell. Proteins are not only produced in the ER, but also controlled. Only proteins that have acquired their correct, biologically active structure are allowed to leave the cell or to be displayed on the cell surface as membrane proteins. Faulty proteins are recognized by dedicated molecular quality control machinery and degraded.
Insights into protein structure formation and its control in the cell are of immediate medical relevance for human pathologies such as Alzheimer’s or Parkinson’s disease – but may also inspire new approaches in medicine and biotechnology. Both of these avenues motivate our research in the laboratory for Cellular Protein Biochemistry (CPB lab). We want to understand principles of protein biogenesis in the cell, understand why these fail in many human diseases, and also provide new biomedical approaches for therapy. In other words: We are driven by curiosity, but also by purpose. To achieve our goals, we use an interdisciplinary approach from biochemistry to mammalian cell biology to understand and ultimately engineer the underlying processes.
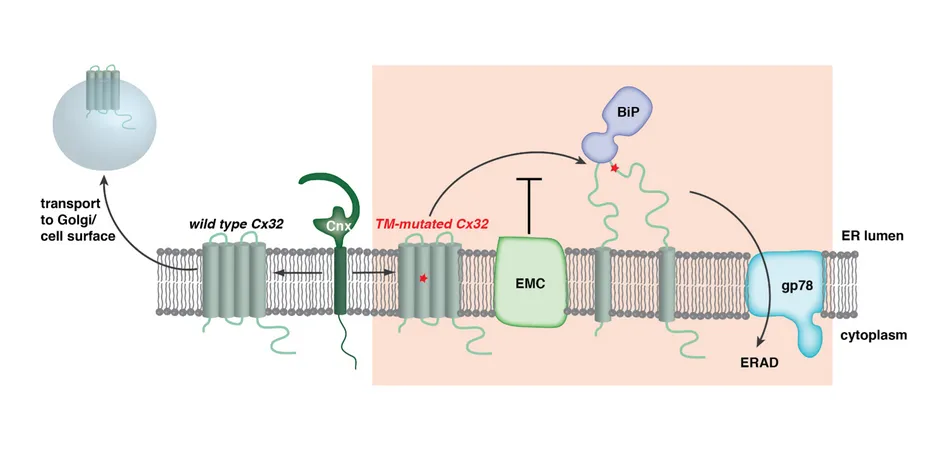
Current work in the CPB lab focuses on two major topics. The first is the quality control of membrane proteins, where principles are still mostly unknown yet have immediate biomedical relevance. Failures in membrane protein biogenesis are associated with a large number of diseases, including neurological disorders. One such example is X-linked Charcot-Marie-Tooth disease, which is caused by mutations in Connexin 32. Charcot-Marie-Tooth disease is the most common genetic disorder of the peripheral nervous system. In our work we were able to show that mutations in Connexin 32 can disturb its proper structure formation and integration into the lipid bilayer [1], a defining step for every membrane protein (Figure 1). This compromises formation of Connexin 32 gap junctions, nano-channels that allow the transport of small molecules within and between cells and are essential in our nervous system.
As a consequence, Schwann cells, which form an insulating layer around nerve cells, die and signal transmission in the nervous system is compromised. Our insights thus provide a molecular explanation for certain types of Charcot-Marie-Tooth disease, which may ultimately help in developing new therapeutic approaches. On the basis of these findings, we have meanwhile significantly expanded our scope and our team working on membrane protein biogenesis. A team of five doctoral candidates within our lab is currently performing systematic analyses on cellular quality control machinery for membrane proteins and on the structural biology of these molecular machines. This team has recently uncovered entirely new mechanisms of how the cell handles misfolded membrane proteins. These insights, some of which will be published shortly, help us in understanding at a molecular level how cells produce a major protein class that allows them to interact and build complex multicellular organisms. Also, the newly identified machines for membrane protein quality that we are analyzing may provide targets for future pharmacological treatment.
Figure 1
The second major research focus of the CPB lab is the biogenesis and engineering of interleukins (ILs), key sigvnaling molecules in our immune system. In humans, more than 40 different ILs exist. They constitute the language of immune cells. Depending on the individual IL, these mount, sustain, or suppress immune reactions in health and disease. Understanding interleukin biogenesis in the cell in more detail can thus provide avenues toward rationally modulating immune responses: dampening immune responses in autoimmunity, but also augmenting them when needed, e.g., to fight cancer wcells. Our work mostly focuses on the interleukin 12 (IL-12) family, which comprises four members (IL-12, IL-23, IL-27 and IL-35, Figure 2A) that span a broad range of biological functions [2] – from activating immune responses to suppressing immunity. As such, IL-12 family members are involved in a large number of human diseases including autoimmune disorders, cancer development, and sepsis [3]. Several studies from our lab in the past five years have revealed the molecular mechanisms of how cells produce IL-12 family members [4-9]. In one of these, in close collaboration with colleagues in the Bavarian NMR Center, TUM Medicine, and the Bioengineering Department at Stanford University, we were able to reveal mechanistic and structural details how cells produce IL-23 [5] (Figure 2B). This study now helps understanding how cells regulate and control protein interactions for signaling molecules (IL-23 consists of two parts), but also provides insights into this pro-inflammatory molecule that already is a successful target of antibody therapies in the clinics. In another study, in close collaboration with TUM Medicine and TUM Physics as well as with immunologists in Paris, we were able to gain detailed insights into how IL-27 acquires its native structure and how cells monitor structure formation of this protein [8]. These studies contribute to the basis of current research in the lab. Here a team of doctoral candidates and postdoctoral researchers is advancing our understanding of hitherto hardly understood IL-12 family members (namely IL-35) and is now using these insights for the rational engineering of interleukins with desired properties: Using these insights, we engineered a new human interleukin, extending the language of human immune cells. Currently we are further developing this rationally engineered biomolecule from our lab as a potential new treatment for sepsis, one of the deadliest diseases in developed countries without any good causal treatment options. Our first patent on the development of this new molecule was awarded with the Innovationspreis 2019 der BioRegionen in Deutschland. Funded by the BMBF and the Else-Kröner-Fresenius Stiftung, we have now built an interdisciplinary team in protein biochemistry, molecular immunology, pharmacology, and medicine, including an expert in the biotech business, to ultimately bring our engineered interleukins to the clinics. Within a few years, we have thus taken the step from basic science to (pre-)clinical translation, which was possible due to the broad range of expertise present at TUM.
Figure 2
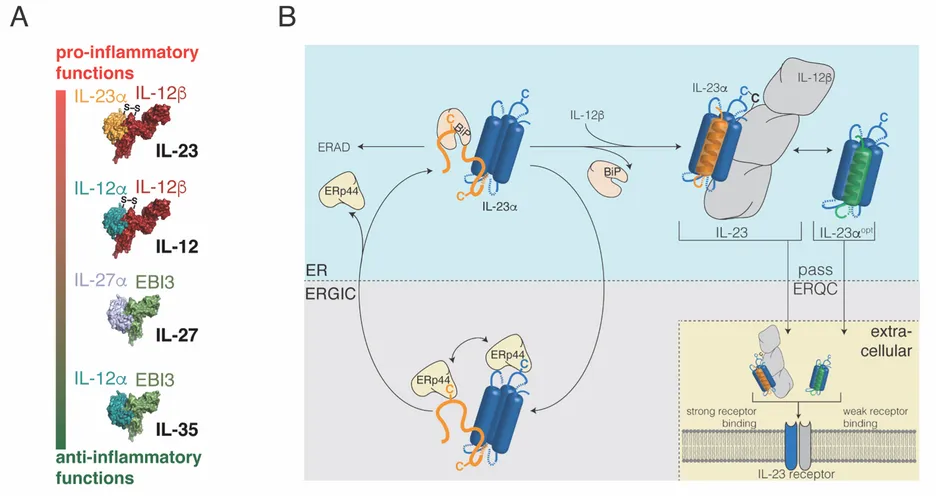
2A) Schematic of the IL-12 family. Each family member is a heterodimer, consisting of one α and one β subunit, and performs distinct roles in regulating immune responses. IL-12 and IL-23 are covalent dimers, connected by a disulfide bond (S-S), whereas IL-27 and IL-35 are non-covalent dimers.
2B) The molecular basis of IL-23 biogenesis. IL-23α is retained in the cell by molecular chaperones that recognize its unstructured first α-helix. Once IL-23α assembles with its partner, IL-12β, it can be secreted and signal via its receptor. In silico optimization of IL-23α allows it to pass ER quality control (ERQC) without IL-12β and act as a signaling molecule on its own. Figure in (B) taken from [5].
[1]
J. Pl. L. Coelho et al., “A network of chaperones prevents and detects failures in membrane protein lipid bilayer integration”, Nature Communications, vol. 10, no. 1, 2019.
[2]
D. A. Vignali and V. K. Kuchroo, “IL-12 family cytokines: immunological playmakers”, Nature Immunology, vol. 13, no. 8, 2012.
[3]
E. D. Tait Wojno, C. A. Hunter and J. S. Stumhofer, “The Immunobiology of the Interleukin-12 Family: Room for Discovery”, Immunity, vol. 50, no. 4, 2019.
[4]
S. Bohnacker et al., “Influence of glycosylation on IL-12 family cytokine biogenesis and function”, Molecular Immunology, vol. 126, 2020.
[5]
S. Meier et al., “The molecular basis of chaperone-mediated interleukin 23 assembly control”, Nature Communications, vol. 10, no. 1, 2019.
[6]
Y. G. Mideksa et al., “Site-Specific Protein Labeling with Fluorophores as a Tool To Monitor Protein Turnover”, ChemBioChem, 2020.
[7]
S. I. Müller, I. Aschenbrenner, M. Zacharias and M. J. Feige, “An Interspecies Analysis Reveals Molecular Construction Principles of Interleukin 27”, Journal of Molecular Biology, vol. 431, no. 12, 2019.
[8]
S. I. Müller et al., “A folding switch regulates interleukin 27 biogenesis and secretion of its alpha-subunit as a cytokine”, Proceedings of the National Academy of Sciences of the United States of America, vol. 116, no. 5, 2019.
[9]
S. Reitberger, P. Haimerl, I. Aschenbrenner, J. Esser-von Bieren and M. J. Feige, “Assembly-induced folding regulates interleukin 12 biogenesis and secretion”, Journal of Biological Chemistry, vol. 292, no. 19, 2017.