Discovering networks in multiple layers of gene regulation
This project explored gene-regulatory networks across different molecular levels. We have developed several computational tools to facilitate the analysis of biological data, enabling the generation of new hypotheses for experimental validation. Our primary objective is to support researchers in analyzing their data more effectively and understanding how biological processes are regulated.


Focus Group: Gene-Regulatory Mechanisms
Dr. Lothar Hennighausen (NIDDK, National Institutes of Health, USA), Alumnus Hans Fischer Senior Fellow, Prof. Priscilla Furth (Georgetown University, USA), Anna Boyksen Fellow | Dr. Markus Hoffmann (TUM, now NIDDK, National Institutes of Health, USA), Doctoral Candidate, now Postdoctoral Fellow | Host: Prof. Markus List (TUM)
Focus Group goal and effort rationale
Multicellular organisms, such as humans for example, comprise a vast number of individual cells. Although each cell contains almost identical DNA, they develop into at least 200 different types, each with its own shape and function. This process relies on various regulatory mechanisms that work at multiple molecular levels. These include genomics (the DNA sequence itself), epigenomics (the structural changes in DNA that control its accessibility), and transcriptomics (the production of RNA from DNA). Controlled adjustments to such regulations are essential for normal processes responding to stimuli, such as pregnancy or immune response to a virus. However, studies have shown that disruptions can lead to diseases such as Alzheimer’s, diabetes, cancer, and autoimmune disorders. In our project, we created new computational tools to investigate these regulatory mechanisms, aiming to support researchers to improve our understanding of how disruptions can cause disease.
NIDDK, National Institutes of Health, and TUM-IAS collaborative research highlights
Inspired by the above goals, our collaboration involving NIDDK, National Institutes of Health, USA, and the TUM-IAS has focused on the following areas:
Regulation in genomics
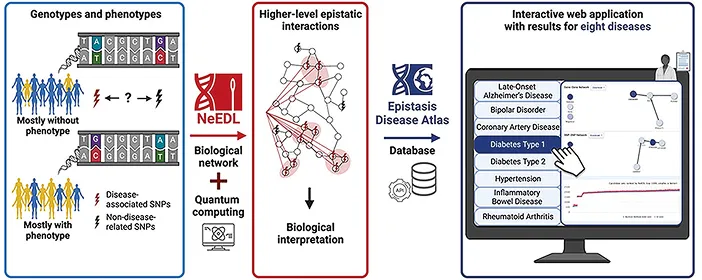
In our study on regulatory networks in genomics, we developed a network-based approach to uncover interactions between single nucleotide genetic variations on the DNA (also called SNPs) that individually have little or no effect but together could contribute to complex diseases. Our tool, NeEDL (network-based epistasis detection via local search), identifies sets of SNPs that could interact with each other and may have an impact on disease risk. Since we know of millions of SNPs, this represents a difficult task. Hence, to make our approach faster, we also explored quantum computing. When tested on diseases, NeEDL showed more robust performance than existing methods, offering new insights into how a combination of genetic interactions may drive disease.
Figure 1
Regulation in epigenomics
To investigate regulatory networks at the epigenomic level, we developed TF-Prioritizer, which identifies proteins called transcription factors (TFs) controlling the production of RNA from the DNA in specific biological conditions. TF-Prioritizer combines data on RNA expression and DNA accessibility to pinpoint TFs that might be important for different physiological conditions. We analyzed gene regulation during pregnancy and lactation in mice, identifying known and new transcription factors that may play important roles in this process. We showed that TF-Prioritizer streamlines complex data analysis, helping researchers better understand the regulation of RNA expression and its links to disease.
Regulation in transcriptomics
We developed two tools to explore how regulatory networks operate within the transcriptomics layer. At this regulatory level, small molecules called microRNAs (miRNAs) help control protein expression by binding to other RNA molecules and degrading them. A single miRNA can interact with many different RNAs, creating a network of connections between them. For instance, if one RNA is very abundant and binds many miRNAs, it can prevent those miRNAs from interacting with other RNAs that they typically regulate. To investigate this network and identify such upregulated RNA-miRNA relationships that could disrupt this regulatory mechanism, we developed the tool spongEffects. We showed that this tool is particularly useful in cancer research, enabling us to detect RNA interactions unique to different breast cancer subtypes and supporting personalized treatment approaches. Building on this, we created circRNA-sponging to examine circularRNAs, which have been shown to accumulate in a cell over time and could, hence, bind many miRNAs, thereby deregulating the network.
Synergistic activities across disciplines during our self-organized international conference: Genetoberfest – crossing bridges between bioinformatics and clinical research (TUM-IAS 2023)
During the Hans Fischer Senior Fellowship, we organized the Genetoberfest conference from October 16 to 19, 2023, at the TUM-IAS building in Garching. The conference drew the attention of 90 participants from all over Europe, the USA, Canada, Qatar, and South Korea. We offered keynote talks, plenary talks, and a wrap-up talk by undergraduate students. We further picked flash talks and two poster sessions from submitted abstracts. During these sessions, we discussed subjects that mattered most for clinical researchers but also for bioinformaticians. Subjects included: (i) the transformative impact of large-scale projects fostering open science, such as The Cancer Genome Atlas or the International Human Epigenome Consortium, (ii) opportunities for using AI in biomedical and clinical research, (iii) limitations in open data sharing and strategies to overcome them, (iv) how high-quality science could and should be incentivized, (v) how software development in biomedical science could be funded, and (vi) what science could learn from applications of AI in industry. The overall goal of our conference was to find common ground between the various disciplines, to determine how we can bridge them, and finally, to advance science.
TUM-IAS doctoral candidate research stay with Dr. Lothar Hennighausen’s group at NIDDK, National Institutes of Health (2023)
In January 2023, the TUM-IAS supported a research visit for its doctoral candidate, Markus Hoffmann, to work with Dr. Lothar Hennighausen’s research group at the NIH's NIDDK in the USA, fostering international collaboration through in-person research. The aim was for Markus Hoffmann to gain direct experience with benchwork methods to better understand the data underlying his biomedical software tool. The visit was so fruitful that Markus Hoffmann chose to stay on as a postdoctoral fellow in Dr. Hennighausen’s lab, enabling him to deepen his biochemical expertise further and apply these insights to his software development.
Selected publications
- Hoffmann, M. et al. Network medicine-based epistasis detection in complex diseases: ready for quantum computing. Nucleic Acids Research 52(17), 10144–10160 (2024).
- Hoffmann, M. et al. TF-Prioritizer: a Java pipeline to prioritize condition-specific transcription factors. GigaScience 12, giad026 (2023).
- Hoffmann, M. et al. spongEffects: ceRNA modules offer patient-specific insights into the miRNA regulatory landscape. Bioinformatics 39(5) (2023).
- Hoffmann, M. et al. circRNA-sponging: a pipeline for extensive analysis of circRNA expression and their role in miRNA sponging. Bioinformatics Advances 3(1), vbad093 (2023).
Full list of publications:
Dr. Lothar Hennighausen
Prof. Priscilla Furth